name: inverse layout: true class: center, middle ,inverse --- # R - Machine Learning Project - Heart disease Diagnosis .footnote[Go directly to [project site](https://github.com/sbme-tutorials/sbe304-fall19-project-sbe304-2021-team09)] --- # Introduction With machine learning, And a simple [dataset](https://archive.ics.uci.edu/ml/datasets/Heart+Disease) of the patient's information, We could accurately detect whether he is diagnosed with heart disease or not. --- layout: false # The dataset Using Cleveland Clinic Foundation dataset ([The processed Version](https://archive.ics.uci.edu/ml/machine-learning-databases/heart-disease/processed.cleveland.data)), Which contains 14 types of data for each of the 302 patients. .left-column[ - **age** - **sex** - **cp** - **trestbps** - **chol** - **fbs** - **restecg** ] .right-column[ - **thalach** - **exang** - **oldpeak** - **slope** - **ca** - **thal** - **num** ] --- #The dataset - **age** - **sex** - **cp** : chest pain type - Value 1: typical angina - Value 2: atypical angina - Value 3: non-anginal pain - Value 4: asymptomatic - **trestbps** : resting blood pressure (in mm Hg on admission to hospital) - **chol** : serum cholestoral in mg/dl - **fbs** : (fasting blood sugar > 120 mg/dl) - 1 = true - 0 = false - **restecg** : resting electrocardiographic results - Value 0: normal - Value 1: having ST-T wave abnormality (T wave inversions and/or ST elevation or depression of > 0.05 mV) - Value 2: showing probable or definite left ventricular hypertrophy by Estes' criteria --- #The dataset - **thalach** : duration of exercise test in minutes - **exang** : exercise induced angina - 1 = yes - 0 = no - **oldpeak** : ST depression induced by exercise relative to rest - **slope** : the slope of the peak exercise ST segment - Value 1: upsloping - Value 2: flat - Value 3: downsloping - **ca** : number of major vessels (0-3) colored by flourosopy - **thal** - 3 = normal - 6 = fixed defect - 7 = reversable defect - **num** : diagnosis of heart disease (angiographic disease status) - Value 0: absent - Value 1-4: present --- template: inverse # Data preprocessing --- # Data preprocessing .left-column[ ###1. Loading the data ] .right-column[ ```sh #specifing the path of the csv file path <- file.path("~","path","to", "processed.cleveland.data") #retrieving dataset from csv file data <- read.csv(path,stringsAsFactors = FALSE) ``` ] --- # Data preprocessing .left-column[ ###1. Loading the data ] .right-column[ ###Output screenshot 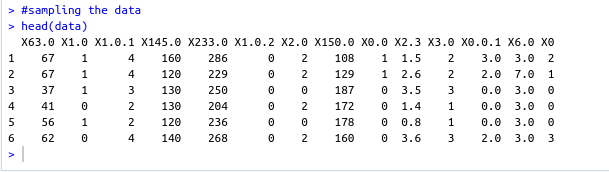 ] --- # Data preprocessing .left-column[ ###1. Loading the data ###2. Adding columns names (Not essential) ] .right-column[ ```sh #data names vector headerNames <- c("age","sex","cp","trestbps","chol" ,"fbs","restecg","thalach","exang","oldpeak", "slope","ca","thal","Class") #renaming the dataframe columns colnames(data) <- headerNames ``` ] --- # Data preprocessing .left-column[ ###1. Loading the data ###2. Adding columns names (Not essential) ] .right-column[ ###Output screenshot 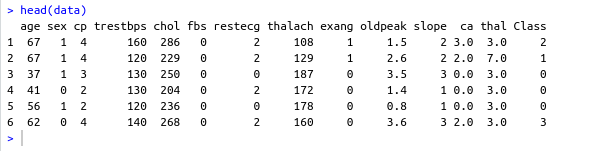 ] --- # Data preprocessing ##3. Maping the class column values to binary values : 'Positive' & 'Negative' ```sh #Factoring won't work with binary nums. #So , we used "Postive" , "Negative" names(data) target <- data$Class makeBinary <- function(x){ if(x == 0){ return ("Negative") } else{ return ("Postive") } } target<-sapply(target,makeBinary) data<-cbind(data,target) #adding new col 'target' to data and removing 'class' keep <- c(names(data)[1:13],names(data[15])) data <- data[keep] ``` --- # Data preprocessing ##3. Maping the class column values to binary values : 'Positive' & 'Negative' ###Output screenshot 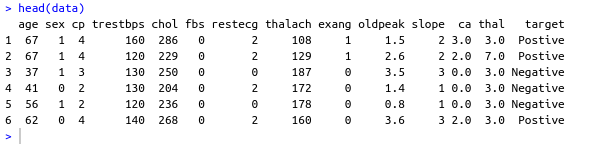 --- # Data preprocessing ##4. Mutating 'ca' & 'thal' data-type to _numeric_ and 'target' to _factor_ ```sh library(dplyr) data <- data %>% mutate( ca = as.numeric(ca), thal = as.numeric(thal), target = as.factor(target) ) ``` --- # Data preprocessing ##4. Mutating 'ca' & 'thal' data-type to _numeric_ and 'target' to _factor_ ###Output screenshot  --- # Data preprocessing ##5. Checking NA values and removing them ```sh sum(is.na(data)) #removing NAs generated by mutate data <- na.omit(data) ``` ###Output screenshot 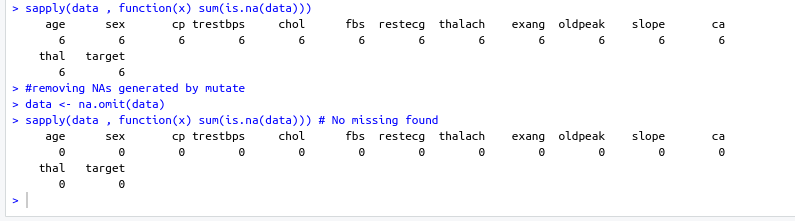 --- template: inverse # Training using [caret](https://cran.r-project.org/web/packages/caret/index.html) --- # Training using [caret](https://cran.r-project.org/web/packages/caret/index.html) ```sh library(caret) cntrl <- trainControl( method ="repeatedcv", repeats = 5, classProbs = TRUE , summaryFunction = twoClassSummary ) ``` --- # Training using [caret](https://cran.r-project.org/web/packages/caret/index.html) ###1. [Naïve Bayes](https://en.wikipedia.org/wiki/Naive_Bayes_classifier#Training) model ```sh set.seed(420) NB_Fit <- train( target ~., data = data, method = "naive_bayes", preProc = c("center","scale"), tuneGrid = expand.grid(.laplace = 0, .usekernel = TRUE, .adjust = 1), trControl = cntrl, metric = "ROC" ) ``` --- # Training using [caret](https://cran.r-project.org/web/packages/caret/index.html) ###1. [Naïve Bayes](https://en.wikipedia.org/wiki/Naive_Bayes_classifier#Training) model ###Output screenshot  --- # Training using [caret](https://cran.r-project.org/web/packages/caret/index.html) ###2. [Decision tree](https://en.wikipedia.org/wiki/Decision_tree_learning) model ```sh DT_Fit <- train( target ~., data = data, method = "rpart", preProc = c("center","scale"), tuneLength =15, trControl = cntrl, metric = "ROC" ) ``` --- # Training using [caret](https://cran.r-project.org/web/packages/caret/index.html) ###2. [ tree](https://en.wikipedia.org/wiki/Decision_tree_learning) model ###Output screenshot 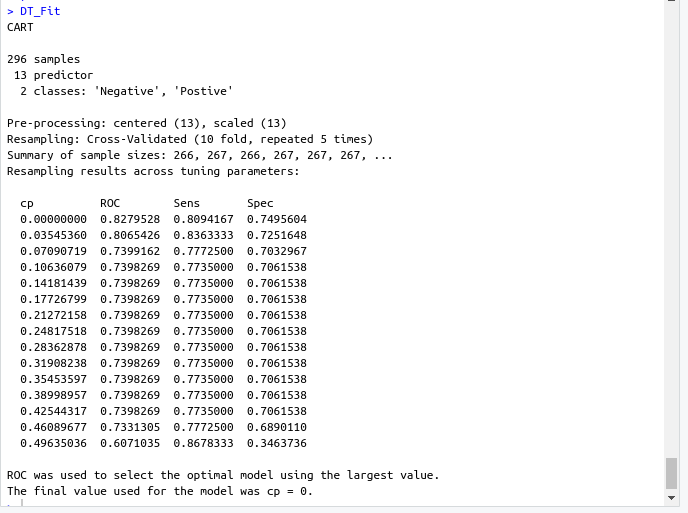 --- # Training using [caret](https://cran.r-project.org/web/packages/caret/index.html) ###2. [Logistic Regression](https://en.wikipedia.org/wiki/Logistic_regression) model ```sh Log_Fit <- train( target ~., data = data, method = "glm", preProc = c("center","scale"), tuneLength =15, trControl = cntrl, metric = "ROC" ) ``` --- # Training using [caret](https://cran.r-project.org/web/packages/caret/index.html) ###2. [Logistic Regression](https://en.wikipedia.org/wiki/Logistic_regression) model ###Output screenshot 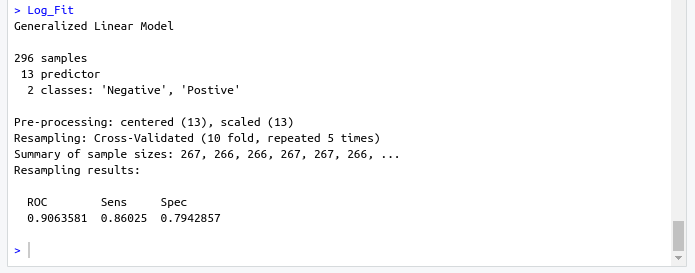 --- # Models summary .left-column[ ###1. Naïve Bayes model ] .right-column[ ```remark - ROC : 0.9055636 - Sens : 0.8755 - Spec : 0.7564835 ``` ] --- # Models summary .left-column[ ###1. Naïve Bayes model ###2. Decision tree model ] .right-column[ ```remark - ROC : 0.9055636 - Sens : 0.8755 - Spec : 0.7564835 ``` ```remark - ROC : 0.8279528 - Sens : 0.8094167 - Spec : 0.7495604 ``` ] --- # Models summary .left-column[ ###1. Naïve Bayes model ###2. Decision tree model ###3. Logistic Regression model ] .right-column[ ```remark - ROC : 0.9055636 - Sens : 0.8755 - Spec : 0.7564835 ``` ```remark - ROC : 0.8279528 - Sens : 0.8094167 - Spec : 0.7495604 ``` ```remark - ROC : 0.9063581 - Sens : 0.86025 - Spec : 0.7942857 ``` ] --- class:center,100%  --- template: inverse class:center, middle #Made by ##[Adel Refat](https://adel-elmala.github.io) --- class:center, middle #THANKS